Starting with the February 28th Training Session, I’ve implemented several exciting new changes and improvements to the Opus23 Basic Certification Training, the day-long training that certifies the student (upon successful completion of the requirements) and permits them to begin using the Opus23 platform on their own clients.
First and foremost, the new sessions will be taught by me, the architect and coder of Opus23. This will allow me to not only bring my own insights into Opus23, but will also allow me to share important elements of my almost four decades of experience in bioinformatics and precision medicine.
With the assumption of the teaching responsibilities, I’ve has coded a new ‘training veneer’ into the Opus23 program. Students work directly inside of Opus23, developing a curation strategy on a shared single client who has a complex health history.

Each student’s curation selections are then saved to a unique data file linked to their account.

This information is monitored by myself and assistants to assess competency by comparing student curation selections to similar curations performed by a panel of seasoned Opus23 professional users. When the student’s curation levels meet the minimum accuracy of the aggregated panel curations, the student essentially ‘passes’ certification.
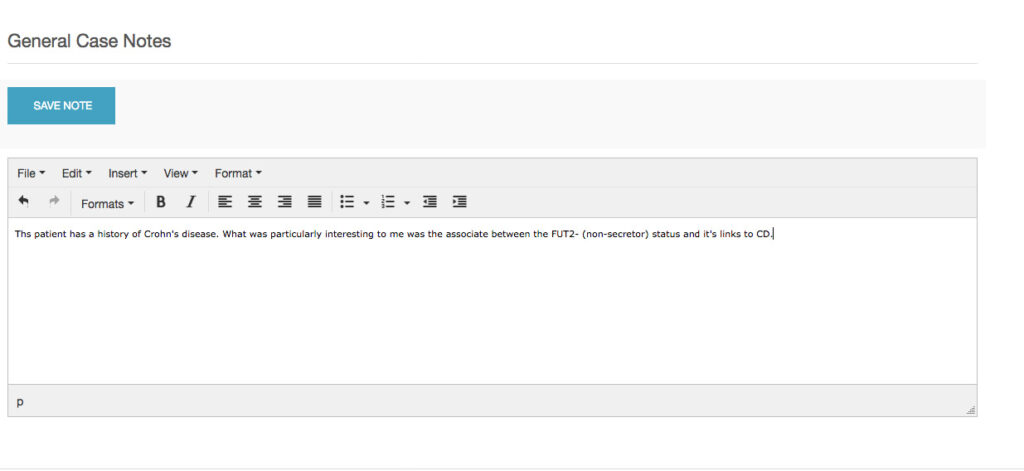
Students provide remarks, observations and justifications using the robust notations functions in Opus23.
This provides for a significant amount of flexibility with regard to working with each student’s unique capabilities. For example, students who quickly master the basic usage of Opus23 genomic apps can be ‘upgraded’ to begin training on the Opus23 microbiome module, Utopia.

The new learning platform also features several new screencast videos of I’ve recorded detailing basic usage and curation. Students can work along with the videos for a true interactive experiences.
Space for these trainings is limited, so if interested, consider reserving your seat now.
Training is limited to licensed health professionals only.
Those who have completed Basic Opus23 Certification can look forward to a series of advanced Opus23 Workshops to begin mid-2021.

1 comment on “Opus23 Certification Training Update”